Selected ATcT [1, 2] enthalpy of formation based on version 1.130 of the Thermochemical Network [3]
This version of ATcT results[4] was generated by additional expansion of version 1.128 [5,6] to include with the calculations provided in reference [4].Methanimine
2D Image:
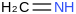
| ΔfH°(0 K) | ΔfH°(298.15 K) | Uncertainty | Units |
|---|---|---|---|
| 96.38 | 88.47 | ± 0.34 | kJ/mol |
3D Image of CH2NH (g)
Top contributors to the provenance of ΔfH° of CH2NH (g)
The 20 contributors listed below account only for 74.2% of the provenance of ΔfH° of CH2NH (g).A total of 102 contributors would be needed to account for 90% of the provenance.
Please note: The list is limited to 20 most important contributors or, if less, a number sufficient to account for 90% of the provenance. The Reference acts as a further link to the relevant references and notes for the measurement. The Measured Quantity is normaly given in the original units; in cases where we have reinterpreted the original measurement, the listed value may differ from that given by the authors. The quoted uncertainty is the a priori uncertainty used as input when constructing the initial Thermochemical Network, and corresponds either to the value proposed by the original authors or to our estimate; if an additional multiplier is given in parentheses immediately after the prior uncertainty, it corresponds to the factor by which the prior uncertainty needed to be multiplied during the ATcT analysis in order to make that particular measurement consistent with the prevailing knowledge contained in the Thermochemical Network.
Top 10 species with enthalpies of formation correlated to the ΔfH° of CH2NH (g)
The correlation coefficient is a number from -1 to 1, with 1 representing perfectly correlated species, -1 representing perfectly anti-correlated species, and 0 representing perfectly uncorrelated species.
| Correlation Coefficent (%) | Species Name | Formula | Image | ΔfH°(0 K) | ΔfH°(298.15 K) | Uncertainty | Units | Relative Molecular Mass | ATcT ID |
|---|---|---|---|---|---|---|---|---|---|
| 37.1 | Methanediimine | HNCNH (g) | 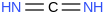 | 153.1 | 146.4 | ± 1.3 | kJ/mol | 42.04006 ± 0.00082 | 151-51-9*0 |
| 23.0 | (1E)-Ethanimine | CH3CHNH (g, trans) | 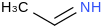 | 55.74 | 39.53 | ± 0.72 | kJ/mol | 43.0678 ± 0.0016 | 56003-81-7*0 |
| 23.0 | Ethanimine | CH3CHNH (g) | 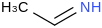 | 55.74 | 41.11 | ± 0.72 | kJ/mol | 43.0678 ± 0.0016 | 20729-41-3*0 |
| 20.9 | Methylimidogen | CH3N (g) | ![C[N]](../images/525.png) | 323.0 | 315.3 | ± 1.0 | kJ/mol | 29.04126 ± 0.00083 | 27770-42-9*0 |
| 19.4 | (1Z)-Ethanimine | CH3CHNH (g, cis) | 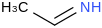 | 58.8 | 44.2 | ± 1.4 | kJ/mol | 43.0678 ± 0.0016 | 56003-82-8*0 |
| 18.8 | N-Methylenemethanamine | CH3NCH2 (g) |  | 94.55 | 79.64 | ± 0.67 | kJ/mol | 43.0678 ± 0.0016 | 1761-67-7*0 |
| 15.6 | Aminomethyl | CH2NH2 (g) | ![[CH2]N](../images/210.png) | 159.40 | 149.56 | ± 0.35 | kJ/mol | 30.04920 ± 0.00085 | 10507-29-6*0 |
| 15.5 | Methylimidogen anion | [CH3N]- (g) | ![C[N-]](../images/1323.png) | 321.1 | 313.4 | ± 1.3 | kJ/mol | 29.04181 ± 0.00083 | 82539-34-2*0 |
| 14.1 | Methyliumylamino | [CH2NH]+ (g) | ![C=[NH+]](../images/1302.png) | 1056.1 | 1049.2 | ± 1.7 | kJ/mol | 29.04071 ± 0.00083 | 65130-65-6*0 |
| 12.6 | Methanediimine cation | [HNCNH]+ (g, cis) | ![N=[C+]=N](../images/1357.png) | 1137.8 | 1133.1 | ± 2.2 | kJ/mol | 42.03951 ± 0.00082 | 1189338-16-6*2 |
Most Influential reactions involving CH2NH (g)
Please note: The list, which is based on a hat (projection) matrix analysis, is limited to no more than 20 largest influences.| 1 |
B. Ruscic, R. E. Pinzon, M. L. Morton, G. von Laszewski, S. Bittner, S. G. Nijsure, K. A. Amin, M. Minkoff, and A. F. Wagner, Introduction to Active Thermochemical Tables: Several "Key" Enthalpies of Formation Revisited. J. Phys. Chem. A 108, 9979-9997 (2004) [DOI: 10.1021/jp047912y] |
|
| 2 |
B. Ruscic, R. E. Pinzon, G. von Laszewski, D. Kodeboyina, A. Burcat, D. Leahy, D. Montoya, and A. F. Wagner, Active Thermochemical Tables: Thermochemistry for the 21st Century. J. Phys. Conf. Ser. 16, 561-570 (2005) [DOI: 10.1088/1742-6596/16/1/078] |
|
| 3 |
B. Ruscic and D. H. Bross, Active Thermochemical Tables (ATcT) values based on ver. 1.130 of the Thermochemical Network. Argonne National Laboratory, Lemont, Illinois 2023; available at ATcT.anl.gov [DOI: 10.17038/CSE/1997229] |
|
| 4 |
N. Genossar, P. B. Changala, B. Gans, J.-C. Loison, S. Hartweg, M.-A. Martin-Drumel, G. A. Garcia, J. F. Stanton, B. Ruscic, and J. H. Baraban Ring-Opening Dynamics of the Cyclopropyl Radical and Cation: the Transition State Nature of the Cyclopropyl Cation J. Am. Chem. Soc. 144, 18518-18525 (2022) [DOI: 10.1021/jacs.2c07740] |
|
| 5 |
B. Ruscic and D. H. Bross Active Thermochemical Tables: The Thermophysical and Thermochemical Properties of Methyl, CH3, and Methylene, CH2, Corrected for Nonrigid Rotor and Anharmonic Oscillator Effects. Mol. Phys. e1969046 (2021) [DOI: 10.1080/00268976.2021.1969046] |
|
| 6 |
J. H. Thorpe, J. L. Kilburn, D. Feller, P. B. Changala, D. H. Bross, B. Ruscic, and J. F. Stanton, Elaborated Thermochemical Treatment of HF, CO, N2, and H2O: Insight into HEAT and Its Extensions J. Chem. Phys. 155, 184109 (2021) [DOI: 10.1063/5.0069322] |
|
| 7 |
B. Ruscic, Uncertainty Quantification in Thermochemistry, Benchmarking Electronic Structure Computations, and Active Thermochemical Tables. Int. J. Quantum Chem. 114, 1097-1101 (2014) [DOI: 10.1002/qua.24605] |
|
| 8 |
B. Ruscic and D. H. Bross, Thermochemistry Computer Aided Chem. Eng. 45, 3-114 (2019) [DOI: 10.1016/B978-0-444-64087-1.00001-2] |
Note that an uncertainty of ± 0.000 kJ/mol indicates that the estimated uncertainty is < ± 0.0005 kJ/mol.
The find function is based on the complete Species Dictionary entries for the appropriate version of the ATcT TN.
The molecule images are rendered by Indigo-depict.
The XYZ renderings are based on Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org/.